Faculty Profile
 Amy K. Sater
Amy K. Sater
Professor and Chair
Department of Biology and Biochemistry
Research Division: Cell and Molecular Biology (Primary), Ecology and Evolution (Joint/Adjunct)
Office: Science & Research 2, 370
Contact: asater@uh.edu - (713) 743-2688
Education: Ph.D., University of Texas at Austin, M.S., Stanford University, B.A., University of California, Santa Cruz
Dr. Amy Sater’s lab seeks to understand the processes by which ectodermal cells become committed to form epidermis, neurons, or glia during embryonic development of the African Clawed Frog, Xenopus laevis. One current project investigates the roles of microRNAs (miRs) in the regulation of ectodermal specification. The lab has identified miRs and miR-targeted RNAs in early neural and epidermal ectoderm and are now testing functions of selected miR-mRNA interactions in the regulation of cell fate.
A second study addresses mechanisms of astrocyte specification and the maintenance of developmental plasticity in astrocytes. Although proliferating neural progenitors acquire the capacity to initiate glial development well after the initiation of neurogenesis, the molecular basis of this “gliogenic switch,” and the paracrine signals and gene regulatory networks that govern the progression from neural progenitor to differentiated astrocyte are poorly understood. The Sater Lab combines genetic, embryological, and genomic approaches to understand the molecular control of astrocyte development.
The Sater Lab has recently begun a new project to develop Xenopus tadpoles as a model system for the investigation of astrocyte responses to traumatic brain injury (TBI). The working hypothesis is that modulation of astrocyte responses to TBI could promote neural regeneration and repair by improving the microenvironment. The lab is currently developing transgenic reporter lines so that astrocyte responses to injury can be monitored in vivo using live imaging. One long-term goal of this project is to use our reporter lines to carry out pharmacological screens for compounds that either reduce the pro-inflammatory response, or promote a neuroprotective, or “pro-regeneration,” response.
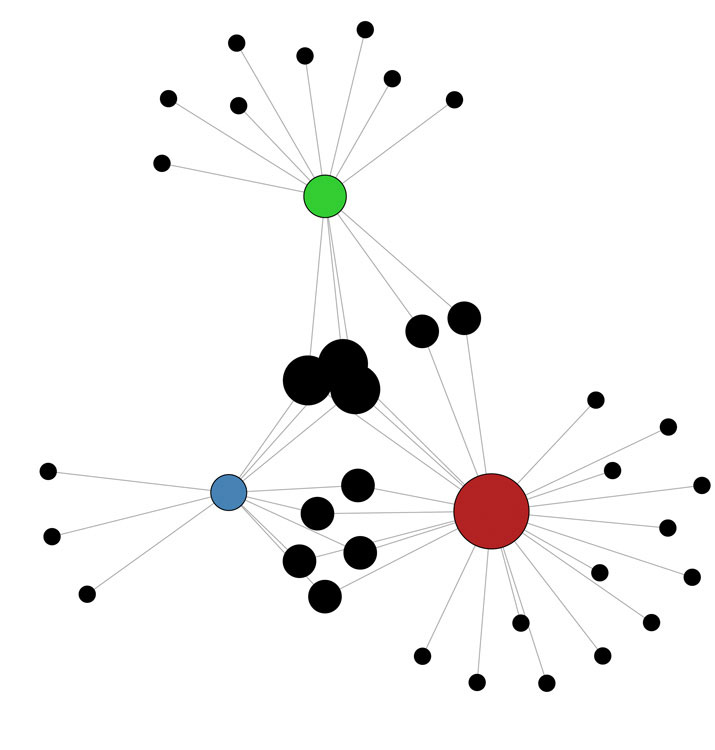
Fig 1. Regulation of the Oct4 orthologues (pou5f3 genes; red, green, blue) by ectodermal microRNAs (black). These genes regulate pluripotency and neural development in Xenopus. This network shows predicted interactions between microRNAs and ectodermal miRs. (from Shah et al., submitted).
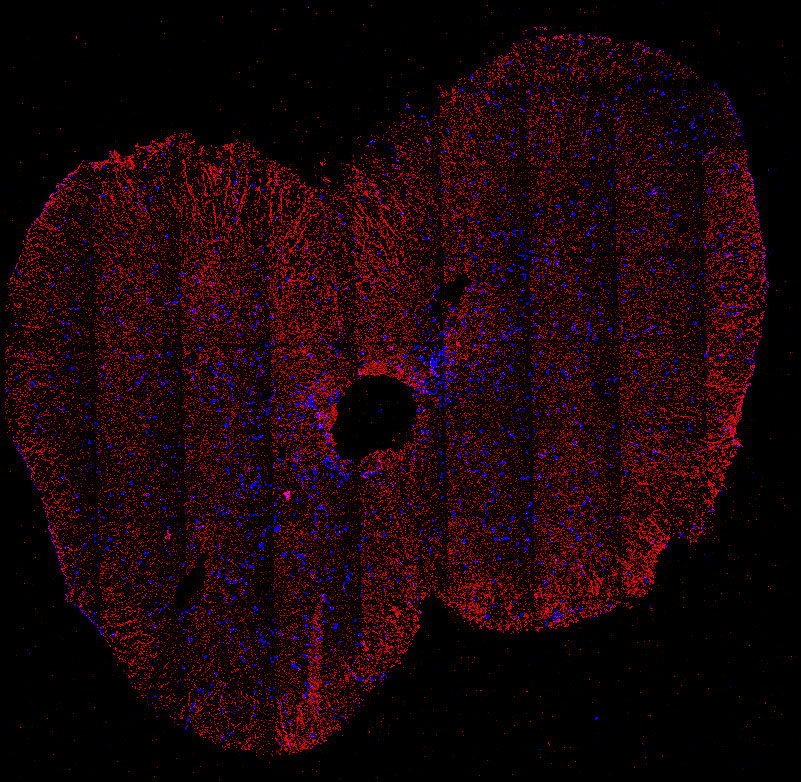
Fig. 2. Tiled immunofluoresence image of the adult cervical spinal cord showing the glutamate transporter GLAST (red) and nuclei (blue). GLAST is expressed in astrocytes and radial glia.

Fig. 3. Xenopus tadpole in mid-metamorphosis.
- Liu C, Lou CH, Shah V, Ritter R, Talley J, Soibam B, Benham A, Zhu H, Perez E, Shieh YE, Gunaratne PH, Sater AK. (2016). Identification of microRNAs and microRNA targets in Xenopus gastrulae: The role of miR-26 in the regulation of Smad1. Dev Biol. 2016 Jan 1;409(1):26-38.
- Shieh, Y-E, Wells, DE, and Sater, AK. (2014). Zygotic expression of Exostosin1 (Ext1) is required for BMP signaling and establishment of dorsal-ventral pattern in Xenopus. Int. J. Dev. Biol. 58: 27 – 34.
- Liu, C, Goswami, M, Talley, J, Chesser-Martinez, PL, Lou, C-H, Sater, AK. (2012). TAK1 promotes BMP4/Smad1 signaling via inhibition of erk MAPK: a new link in the FGF/BMP regulatory network. Differentiation, 83(4):210-219.
- Wells DE†, Gutierrez L, Xu Z, Krylov V, Macha J, Blankenburg KP, Hitchens M, Bellot LJ, Spivey M, Stemple DL, Kowis A, Ye Y, Pasternak S, Owen J, Tran T, Slavikova R, Tumova L, Tlapakova T, Seifertova E, Scherer SE, Sater AK. (2011). A genetic map of Xenopus tropicalis. Dev Biol. 2011 Jun 1;354(1):1-8.
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science. 2010 Apr 30;328(5978):633-6.
- Book Chapter: “The Evolution of Gene Regulation” for D. Graur, Molecular and Genome Evolution, Sinauer Associates. Published Dec 2015.
Xenopus Genome Steering Committee, 2005-2013
Instructor, Cold Spring Harbor Laboratories “Cell and Developmental Biology of Xenopus” course, 2011-2014
Adjunct member, Graduate Faculty, Graduate School of Biological Sciences, University of Texas Health Science Center at Houston, Fall 2007 – present
Member, Genetics Society of America (GSA)
Member, Society for Developmental Biology (SDB)